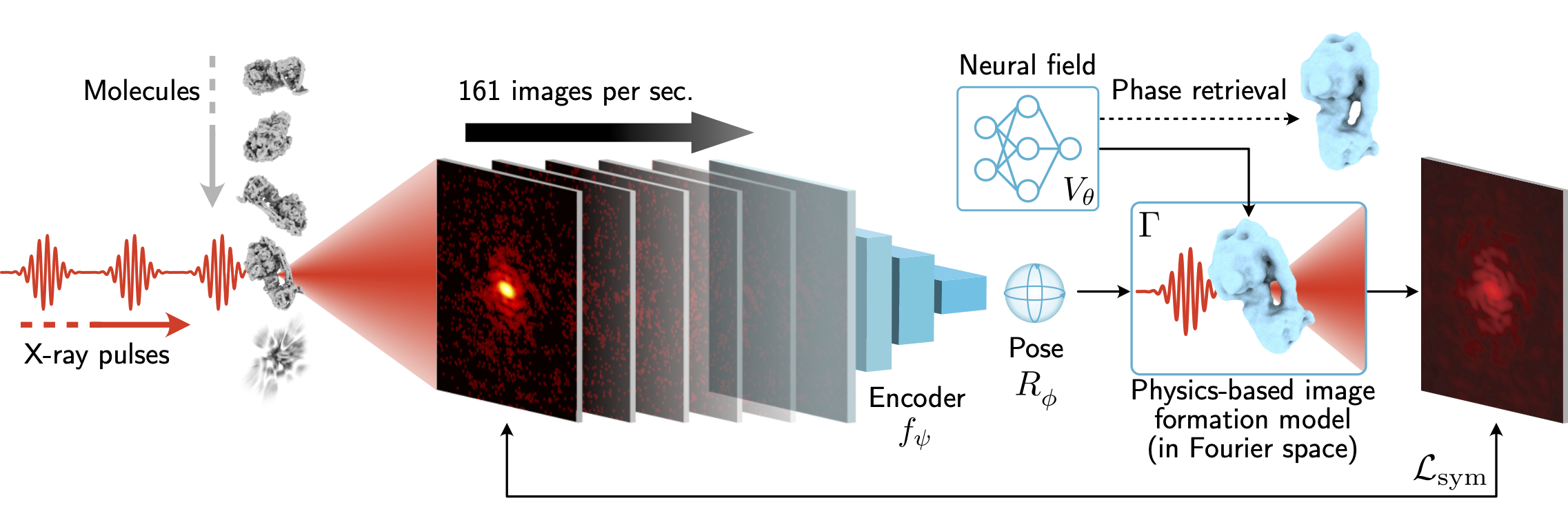
X-ray free-electron lasers (XFELs) offer unique capabilities for measuring the structure and dynamics of biomolecules, helping us understand the basic building blocks of life. Notably, high-repetition-rate XFELs enable single particle imaging (X-ray SPI) where individual, weakly scattering biomolecules are imaged under near-physiological conditions with the opportunity to access fleeting states that cannot be captured in cryogenic or crystallized conditions. Existing X-ray SPI reconstruction algorithms, which estimate the particle orientation for each image independently, are slow and memory-intensive when handling the massive datasets generated by emerging XFELs. Here, we introduce X-RAI (X-Ray SPI with Amortized Inference), an online reconstruction framework that estimates the structure of a 3D macromolecule from large X-ray SPI datasets. X-RAI consists of a convolutional encoder, which amortizes pose estimation over large datasets, as well as a physics-based decoder, which employs an implicit neural representation to enable high-quality 3D reconstruction in an end-to-end, self-supervised manner. We demonstrate that X-RAI achieves state-of-the-art performance for small-scale datasets in simulation and challenging experimental settings and demonstrate its unprecedented ability to process large datasets containing millions of diffraction images in an online fashion. These abilities signify a paradigm shift in X-ray SPI towards real-time reconstruction.

Comparison of X-RAI with M-TIP on reconstructing datasets with 50,000 images in an offline setting. Each row corresponds to a separate dataset simulated using Skopi with the protein data bank (PDB) code specified in the leftmost column. For both datasets, X-RAI is able to reconstruct each particle to within 4 nanometers of resolution, exceeding the quality of M-TIP. For the protein 6J5I, M-TIP fails to reconstruct the intensity volume, resulting in the degenerate density volume shown.

Online reconstruction of the 50S ribosomal subunit (PDB: 5O60). Remarkably, X-RAI is able to reconstruct the protein even when acquiring the data sequentially in batches of 64 images. Two synthetic datasets with varying levels of beam fluence (photons per pulse) are shown in order to illustrate the effect of signal level on convergence speed. Our method converges to a resolution of about 1.7 nm after processing 5 million images when handling images with 1e13 photons per pulse. On the other hand, when operating under a lower fluence level of 6e12 photons per pulse, X-RAI only converges to a resolution of 1.9 nm, demonstrating the effect of fluence on resolution. Since our method processes data serially, it can help experimentalists decide when to cease data collection via the results from live reconstruction.

Reconstruction of the gold nanoparticle from experimental data collected at the European XFEL. To enforce the known cubic symmetry of the virus, we augment the encoder's orientation estimates with the symmetry rotations of the cube, effectively spreading out the pose estimates over SO(3). The resulting phased density reconstruction is 4.56 nm in resolution. The diffraction patterns are displayed with reduced contrast.
An overview of the capabilities and opportunities of X-ray single particle imaging.
Review articles on X-ray crystallography and cryo-electron microscopy, the leading methods for protein structure determination.
CryoAI: Amortized Inference of Poses for Ab Initio Reconstruction of 3D Molecular Volumes from Real Cryo-EM Images
An autoencoder architecture for amortized pose estimation in homogeneous cryo-EM reconstruction.
CryoDRGN-AI
Ab initio heterogeneous reconstruction for cryo-EM and cryo-ET.
@article{shenoy2025xrai,
author = {Shenoy, Jay and Levy, Axel and Ayyer, Kartik and Poitevin, Frédéric and Wetzstein, Gordon},
title = {Scalable 3D reconstruction for X-ray single particle imaging with online machine learning},
journal = {Nature Communications},
year = {2025},
doi = {10.1038/s41467-025-62226-7}
}